Learn Biacore from Publications
Measuring the equilibrium dissociation constant (KD) of biomolecules with very fast binding on-rates and off-rates using a Biacore system often involves performing experiments at a series of suitable concentrations of the ligand. The crucial point is to allow the binding to reach equilibrium levels before starting the dissociation and then plot the equilibrium binding responses to determine the KD. See an example in the publication shown binding wasn’t reach equilibrium level in some of concentrations.
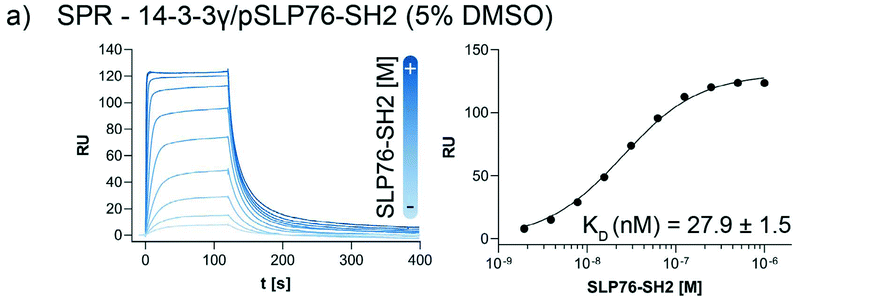
Fig.1 Assay set-up for the 14-3-3γ/SLP76 PPI. a) SPR binding assay of the SLP76-SH2 construct binding to 14-3-3γ in the presence of 5% DMSO. The KD value has been extrapolated from the binding curve reported on the right of the sensorgrams. (Publication title as below and link)
Soini L, Redhead M, Westwood M, Leysen S, Davis J, Ottmann C. Identification of molecular glues of the SLP76/14-3-3 protein-protein interaction. RSC Med Chem. 2021 Aug 2;12(9):1555-1564. doi: 10.1039/d1md00172h. PMID: 34667951; PMCID: PMC8459327.
Kinetics Exhibit Very Fast Binding on/off-Rates
The concentrations of the biomolecules can significantly impact the accuracy and reliability of the binding kinetics measured using a Biacore system. It is crucial to operate at concentrations above the equilibrium concentration. Here are some key considerations:
- Optimize Ligand Immobilization: Begin by optimizing the immobilization of the ligand on the sensor surface. Ensure stable and reproducible immobilization to provide a consistent platform for experiments.
- Determine Suitable Concentrations: Conduct preliminary experiments to determine a suitable range of concentrations for the ligand. Aim for concentrations that provide a measurable response within the dynamic range of the instrument.
- Association and Equilibration: Inject the analyte at different concentrations over the ligand-coated surface to initiate the association phase. Allow the system to equilibrate, ensuring that the binding reaches a steady state or equilibrium before initiating the dissociation phase.
- Dissociation Phase: Initiate the dissociation phase after reaching equilibrium. Monitor the dissociation of the analyte from the ligand-bound surface.
- Concentration Series: Repeat the association and dissociation steps for a concentration series of the analyte. Ensure that the chosen concentrations cover a range that allows for accurate determination of the equilibrium dissociation constant (KD).
- Control for Mass Transport Limitations: Address potential mass transport limitations by optimizing the injection and flow conditions. Adequate mass transport is crucial for accurately measuring fast kinetics.
- Fit to Binding Models: Fit the data to appropriate binding models that account for fast kinetics. Consider models such as 1:1 binding, bivalent binding, or more complex models, depending on the characteristics of the interaction.
- Quality Controls and Statistical Analysis: Implement quality controls, including reference surfaces and negative controls, to ensure specificity. Perform statistical analyses to validate the reliability of the obtained equilibrium kinetics.
- Data Visualization: Visualize the equilibrium kinetics data by plotting curves that represent the association and dissociation phases at different concentrations. This provides a comprehensive view of the binding kinetics at equilibrium.
- Interpretation: Interpret the obtained equilibrium kinetics to gain insights into the binding affinity and kinetics of the biomolecular interaction. Consistent and reproducible data across experiments enhance confidence in the obtained results.
Given the complexity of the SPR technology, seeking professional assistance is highly advisable before Biacore experiment and publishing the results.
How His-tagged proteins deployed in Biacore SPR
Biacore and SPR technology in Applications

