Learn Biacore from Publications
Baseline drifting in Biacore analysis refers to the slow, gradual shift in the baseline SPR signal that occurs over time during a biomolecular interaction analysis experiment. It is a common issue that can affect the accuracy and precision of the data obtained from a Biacore instrument. See an example in the following publication.
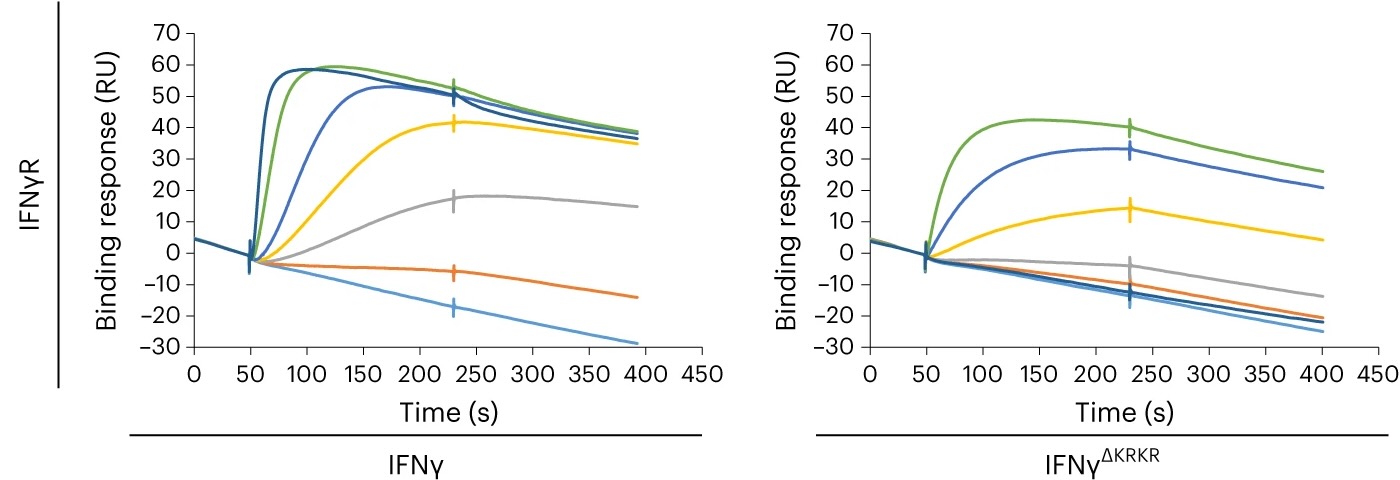
Fig.1g, IFNγ (left) or IFNγΔKRKR (right) was injected over an HS-activated surface (top) or an IFNγR1-activated surface (bottom) over 180 s, and the binding response in resonance units (RU) was recorded as a function of time. Each set of sensorgrams was obtained with IFNγ at (from bottom to top) 0, 25, 50, 75, 100, 150, 200 and 500 nM for the HS surface and at 0, 1, 2.5, 5, 10, 25 and 50 nM for the IFNγR1 surface. (Publication title as below and link)
Kemna J, Gout E, Daniau L, Lao J, Weißert K, Ammann S, Kühn R, Richter M, Molenda C, Sporbert A, Zocholl D, Klopfleisch R, Lortat-Jacob H, Aichele P, Kammertoens T, Blankenstein T. IFNγ binding to extracellular matrix prevents fatal systemic toxicity. Nat Immunol. 2023 Mar;24(3):414-422. doi: 10.1038/s41590-023-01420-5. Epub 2023 Feb 2. Erratum in: Nat Immunol. 2023 Feb 22;: PMID: 36732425; PMCID: PMC9977683.
What Cause Baseline Drifting in Biacore Analysis?
- Baseline drifting can be caused by a variety of factors, including temperature changes, sample evaporation, buffer changes, and changes in the refractive index of the buffer or sample. The drifting baseline signal can be caused by changes in the SPR signal that are not related to the binding interaction being studied, making it difficult to distinguish between real binding events and noise.
- Correcting baseline drift using digital calculations or computing programs can be an effective way to reduce the effects of drift and improve the accuracy of Biacore analysis results. However, it is important to note that these correction methods are based on assumptions and models of the system and may not be completely accurate.
- Baseline drifting can occur in Biacore analysis regardless of whether ligands are directly immobilized on the sensor chip or captured onto the surface.
- However, the extent of baseline drift may depend on the specific immobilization method used, as well as the properties of the ligand and receptor being studied. For example, if the ligand is prone to non-specific binding or aggregation, this may increase the extent of baseline drift and make it more difficult to correct.
How to Prevent or Minimize Baseline Drifting
By taking these steps, it is possible to minimize the effects of baseline drifting and improve the accuracy and precision of Biacore analysis.
- Use stable buffer conditions: Use a buffer system that is stable over time and consistent between experiments, such as low salt concentration buffers or temperature-controlled systems.
- Control temperature: Keep the temperature of the system stable throughout the experiment.
- Maintain constant sample volume: Maintain a constant sample volume throughout the experiment to prevent sample evaporation.
- Use reference surface: Use a reference surface to correct for baseline drifting caused by changes in the refractive index of the buffer or sample.
- Use control sample: Use a control sample, such as a buffer blank, to monitor baseline drift and correct for changes that are not related to the biomolecular interaction being studied.
- Run buffer injections: Injecting buffer between each sample injection can help to stabilize the system and correct for baseline drift.
In summary, while baseline drifting can be a challenge in Biacore analysis, the results can still be trusted if appropriate correction methods are used. It is important to carefully monitor and correct for baseline drift in order to obtain accurate and reliable results.
How His-tagged proteins deployed in Biacore SPR
Biacore and SPR technology in Applications

